predicting recurrence in patients with brain metastases
Read the paper here: LINK
This past week I had my first first-author publication published. It’s satisfying to see a project I put a lot of thought and effort into come to something tangible. This paper is the combination of a cool technique applied to a unique dataset.
The technique is radiomics, a method for extracting quantitative features from medical imaging datasets and developing predictive models with those features. The dataset is a cohort of patients who develop brain metastases, have them resected, and then receive stereotactic radiosurgery to the surgical cavity. Our question is whether or not radiomics features could be used to predict which patients will experience a recurrence within the resection cavity. This is what the MRIs look like:
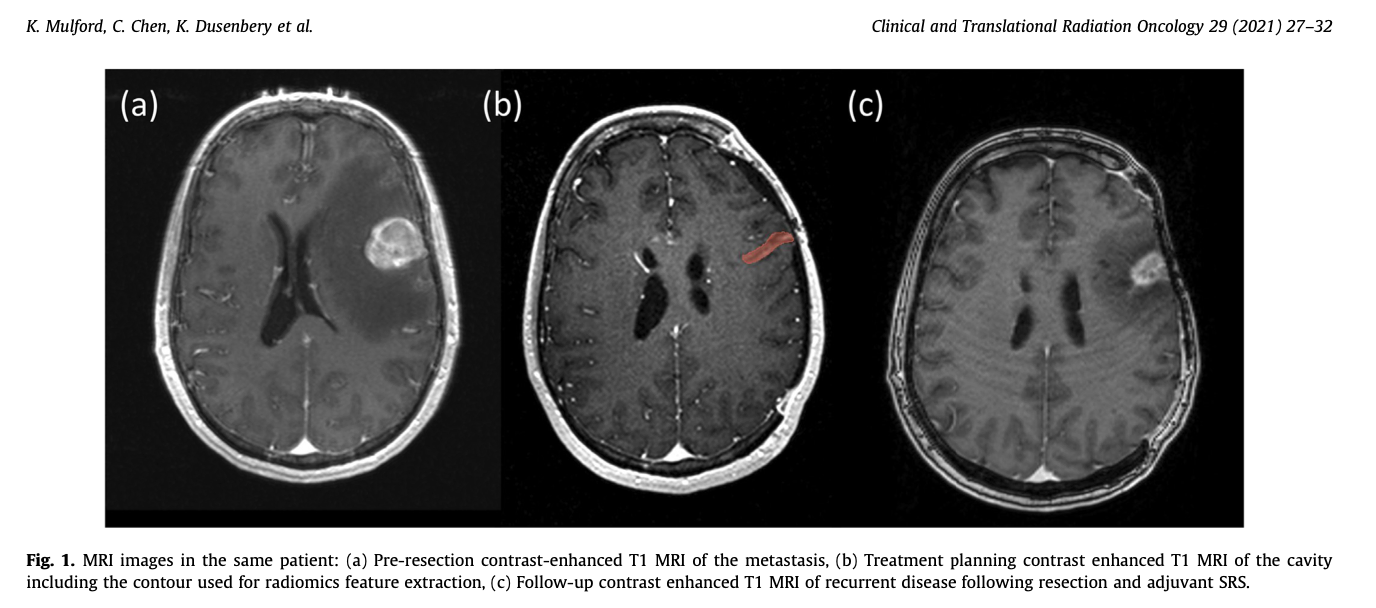
If you don’t want to read the rest of the article - our conclusion was that a radiomics based predictive model does moderately succesfull at identifying patients who will eventually experience a local recurrence and substantiallyt outperforms clinically derived features alone. Our hunch is that more data would increase this effect.
The Process
The first step is the assembly of the dataset - we were looking for patients who had
- Developed a brain metastasis secondary to a cancer somewhere else in the body
- A surgical resection (removal) of that brain metastasis
- Adjuvant stereotactic radiosurgery to the resection cavity
This identified 73 cavities from 69 patients. Not a lot for machine learning, but it is what it is.
Next the MRIs were processed to have a uniform signal level by way of batched histogram matching. The resection cavities were contoured for feature extraction and ~100 shape, intensity, and texture features were extracted from each cavity region of interest (for more info on radiomics feature extraction you can look through this site: Link).
Now the fun part - model building! With ~100 features and 73 instances, some feature selection will be necessary. I cannot tell you how many papers in top-ranked journals I’ve seen make the mistake of performing feature selection before model training, which leaks validation data information into the model training phase!
Within our cross-validation scheme (K-fold with K=5) I performed recursive-feature-elimination on a logistic regression screener. The number of features was limited to prevent overfitting - we ended up with 6 features. Then those features were used to train a gradient-boosting classifier. Average performance over the K validation folds was reported.
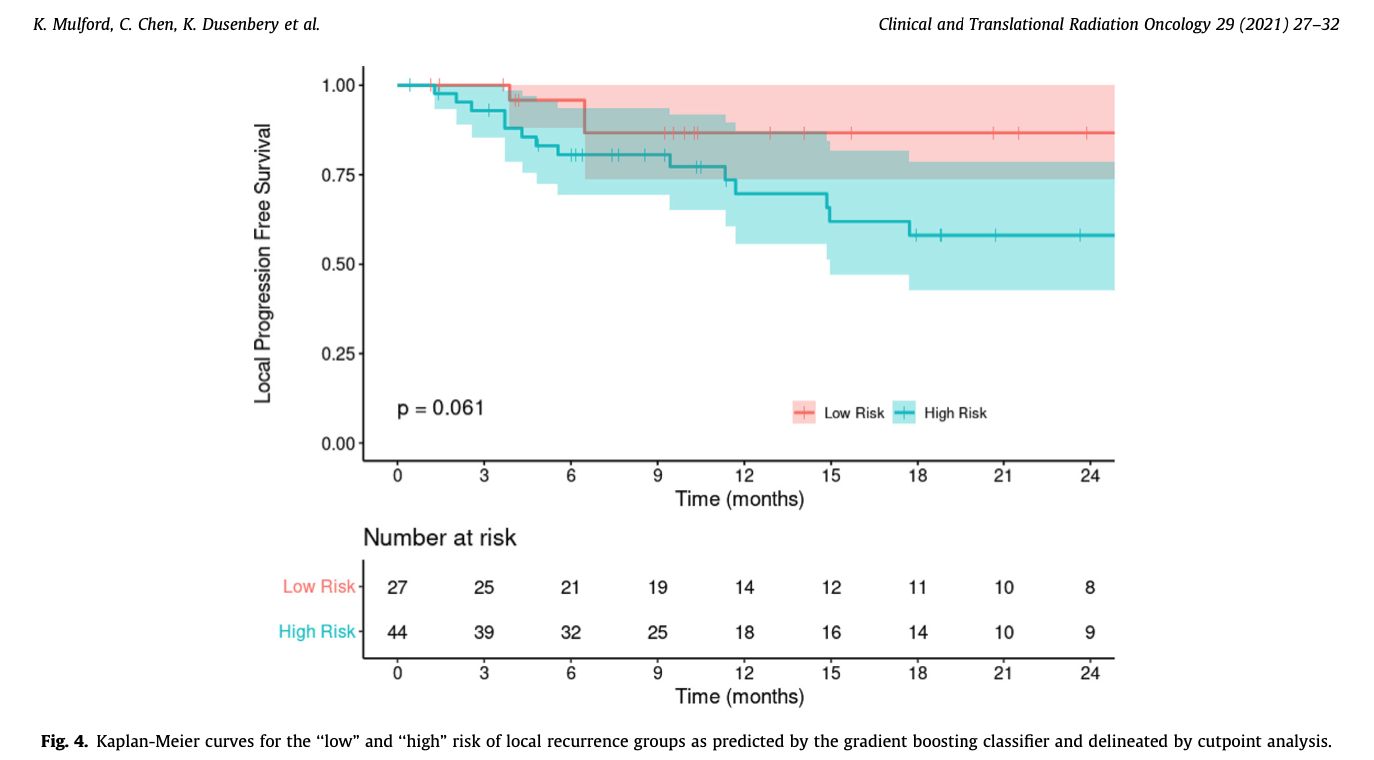
The last step was to take the probability output of the GBM classifier and identify a cutpoint between “low” and “high” risk groups that maximized the sum of sensitivity and specificity between them, and then to compare the “local recurrence” survival curves between the two groups.
Results
You can read the article for all the details - so I will keep this brief.
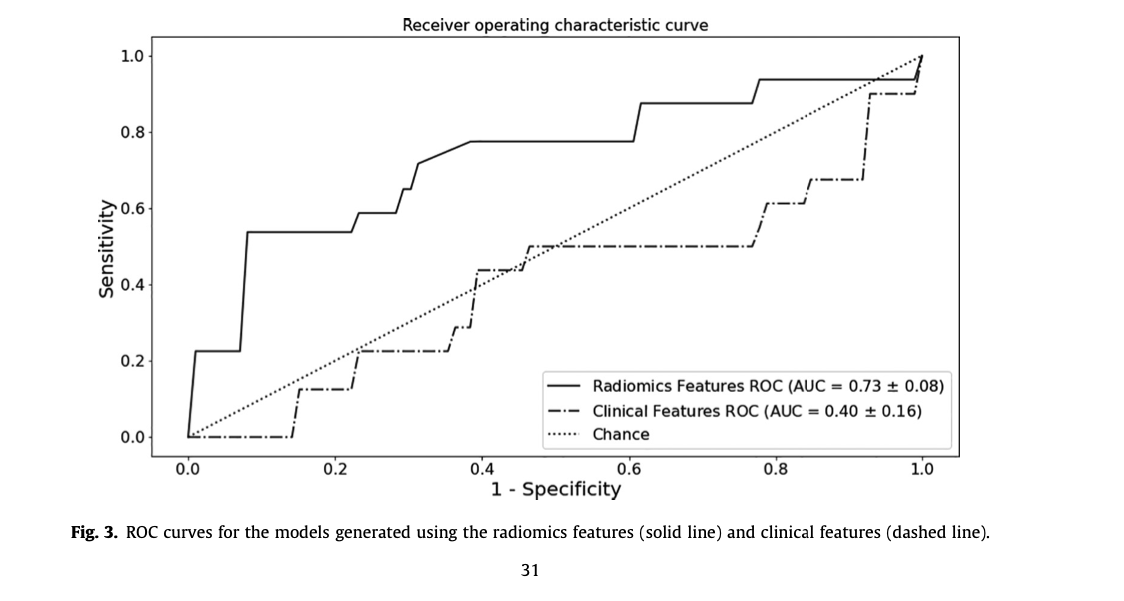
- Our image-feature based model gives us an AUC of 0.73, with an accuracy of 76%. This outperforms the model trained on the clinical features alone (see figure).
- The cutpoint analysis on the GBM class propbabilities shows that patients placed into the “high risk” category by the model have worse outcomes. While this does not rise to the 0.05 level of statistical significance - the trend is clearly evident.
Conclusions
The big conclusion is that radiomics models not only apply to solid tumors, but also to the resection cavities left behind - this could potentially lead the way towards personalized adjuvant treatment planning schemes. Unfortunately, the opportunities to get more data for this project are small - as the department upgraded its GammaKnife and they now do a hypofractionated scheme for adjuvant therapy to cavities.
Overall this was one of the first projects I took on as a graduate student, and it was really cool to see it out in the world. Thanks for reading!